Class 12 Biology Chapter 6 Important Questions Molecular Basis of Inheritance
Question 1.
Write the dual purpose served by deoxyribonucleoside triphosphates in polymerisation. (2018)
Answer:
Deoxyribonucleoside Triphosphates (dNTPs) serve the dual purpose of
- acting as a substrate.
- providing energy (from two terminal phosphates).
Question 2.
Why does hnRNA undergo splicing? Where does splicing occur in the cell? (Delhi 2015C)
Answer:
hnRNA undergoes splicing to remove non-coding sequences, i.e. introns and joins exons. Splicing occurs in the nucleus of the cell.
Question 3.
Name the enzyme that transcribes hnRNA in eukaryotes. (Delhi 2015C)
Or
Write the function of RNA polymerase-II. (Foreign 2015)
Answer:
The RNA polymerase-II transcribes the precursor of mRNA, i.e. the heterogeneous nuclear RNA (hnRNA).
Question 4.
Why is RNA more reactive in comparison to DNA? (Delhi 2015C)
Answer:
The OH-group in the ribose of RNA at 3’end makes the molecule more reactive, compared with DNA. RNA is also not stable under alkaline conditions.
Question 5.
Name the negatively charged and positively charged components of a nucleosome. (Delhi 2015C)
Or
Name the positively charged protein around which the negatively charged DNA wrapped. (All India 2012C)
Answer:
Histone proteins forming an octamer is the positively charged component and the DNA helix is the negatively charged component of nucleosome.
Question 6.
What is cistron? (All India 2015)
Answer:
The segment of DNA coding for a polypeptide is known as cistron.
Question 7.
Name the transcriptionally active region of chromatin in a nucleus. (Delhi 2015)
Answer:
Euchromatin (lightly stained) is the transcriptionally active region of chromatin in a nucleus.
Question 8.
What will happen if DNA replication is not followed by cell division in a eukaryotic cell? (All India 2014C)
Answer:
If cell division is not followed after DNA replication then replicated chromosomes (DNA) would not be distributed to daughter nuclei. A repeated replication of DNA without any cell division results in the accumulation of DNA inside the cell.
This would increase the volume of the cell nucleus, thereby causing cell expansion. Further, it will lead to polyploidy.
Question 9.
Why is it not possible for an alien DNA to become part of chromosome anywhere along its length and replicate normally? (All India 2014)
Answer:
An alien DNA cannot become a part of chromosome anywhere along its length and replicate normally due to the absence of origin of replication where the replication process is initiated.
Question 10.
Name the enzyme and state its property that is responsible for continuous and discontinuous replication of the two strands of a DNA molecule. (Delhi 2013)
Answer:
DNA-dependent DNA polymerase This enzyme can polymerise deoxynucleotides in 5′ → 3′ direction only.
Due to this, replication of DNA is continuous in one strand with polarity 3′ → 5’while discontinuous in another polarity 5′ → 3′.
Question 11.
Name the enzyme that joins the small fragment of DNA of a lagging strand during DNA replication. (Delhi 2013C)
Answer:
DNAligase.
Question 12.
Which one of an intron and an exon is the reminiscent of antiquity? (All India 2013C)
Answer:
Intron is considered to be the reminiscent of antiquity because these are non-coding sequences. (1)
Question 13.
Name the specific components and the linkage between them that forms deoxyadenosine. (Delhi 2013C)
Answer:
The specific components of deoxyadenosine are adenine and deoxyribose. These are linked by N-glycosidic linkage.
Question 14.
Which one out of rho factor and sigma factor acts as an initiation factor during transcription in a prokaryote? (Delhi 2013C)
Answer:
Sigma factor acts as an initiation factor and binds to the promoter during transcription in prokaryotes.
Question 15.
Name the enzyme involved in continuous replication of DNA strand. Mention the polarity of the template strand. (All India 2012)
Answer:
Enzyme involved in continuous replication of DNA strand is DNA polymerase.
Template strand has 3′ → 5′ polarity.
Question 16.
Name the two basic amino acids that provide positive charges to histone proteins. (Delhi 2012C)
Answer:
Lysine and arginine.
Question 17.
If the base adenine constitutes 31% of an isolated DNA fragment, then what is the expected percentage of the base cytosine in it? (Delhi 2011c)
Answer:
According to Chargaff’s rule A + G = C+ T = 50%
∴ if A = 31% then T = 31%;
C + T = 50%
∴ C = 50% – 31% = 19%
Question 18.
A structural gene has two DNA strands X and Y shown along side. Identify the template strand. (All India 2010C)
Answer:
‘X’ is template strand. It is because the template strand has the polarity in 3′ → 5′ direction.
Question 19.
What is an origin of replication in a chromosome? State its function. (Delhi 2019)
Answer:
Replication does not initiate randomly at any place in DNA (chromosome). So, there is a definite region in DNA (chromosome) termed as origin of replication. It is the place from where DNA replication originates. It helps in propagation of a piece of DNA, during recombinant DNA procedures.
Question 20.
Although a prokaryotic cell has no defined nucleus, yet DNA is not scattered throughout the cell. Explain. (2018)
Answer:
Prokaryotes lack a defined nucleus. However, the DNA is not scattered throughout the cell. This is due to the fact that the negatively charged DNA is held with some positively charged proteins in nucleoid or the middle region. In this region, the DNA is organised in large loops held by proteins.
Question 21.
Describe the structure of a nucleosome. (Delhi 2017)
Or
Draw a labelled diagram^of a nucleosome. Where is it found in a cell? (Foreign 2014; All India 2012)
Or
How do histones acquire positive charge? Delhi 2011
Answer:
Nucleosome is a complex of negatively charged DNA. wrapped around the positively charged histone octamer (unit of 8 molecules of histone). It is found in the nucleus of the cell.
It is made up of four types of proteins; H2A, H2B, H3 and H4 occurring in pairs. The histone proteins acquire positive charge due to abundance of amino acid residues, i.e. lysine and arginines. Both these amino acids carry positive charges in their side chains. A typical nucleosome consists of 200 base pairs of DNA helix.
Question 22.
Discuss the role enzyme DNA ligase plays during DNA replication. (All India 2016)
Answer:
DNA ligase facilitates the joining of Okazaki fragments in lagging DNA strands together by catalysing the formation Of phosphodiester bond. It also plays a role in repairing single-strand breaks in duplex DNA.
Question 23.
In a typical nucleus, some regions of chromatin are stained light and others dark. Explain why is it so and what is its significance. (Outside Delhi 2016C)
Answer:
In a typical nucleus, some regions of chromatin are stained light because of loose packing of chromatin and some regions of chromatin are stained dark because, chromatin is densely packed. Euchromatin is transcriptionally active chromatin (lightly stained) while heterochromatin (darkly stained) is transcriptionally inactive.
Question 24.
Explain the two factors responsible for conferring stability to double helix structure of DNA. (All India 2014)
Answer:
Two factors responsible for conferring stability to double helix structure of DNA are
- Stacking of one base pair over other.
- H-bonds between nitrogenous bases.
Question 25.
State the difference between the structural genes in a transcription unit of prokaryotes and eukaryotes. (All India 2014)
Answer:
Prokaryotic structural genes are continuous and are without any non-coding region, while eukaryotic structural genes contain exons (coding DNA) and introns (non-coding DNA).
Question 26.
Show DNA replication with the help of a diagram only. (All India 2014C; Delhi 2012)
Answer:
The replication fork of DNA formed during DNA replication.
Question 27.
A template strand is given below. Write down the corresponding coding strand and the mRNA strand that can be formed, along with their polarity. (Foreign; 2014)
3′- ATGCATGCATGCATGCA TGCATGC-5′
Answer:
For the given template strand 3- ATGCATGCAT GCATGCATGCATGC- 5′
Coding strand is 5′- TACGTACGTACGTACGTACG TACG – 3′
and mRNA strand is 5′- UACGUACGUACGUACGU ACGUACG – 3′
Question 28.
Draw a schematic diagram of a part of double-stranded dinucleotide DNA chain having all the four nitrogenous bases showing the correct polarity. (Delhi 2012)
Answer:
Schematic diagram of a double stranded dinucleotide DNA chain having all the four nitrogenous bases (A, T, G, C) with polarity.
Question 29.
State the functions of the following in a prokaryote:
(i) tRNA
(ii) rRNA (All India 2012)
Answer:
(i) tRNA helps in transferring amino acids to ribosome for the synthesis of polypeptide chain.
(ii) rRNA plays structural and catalytic role during translation.
Question 30.
Differentiate between a cistron and an exon. (All India 2012C)
Answer:
Differences between cistron and exon are as follows
| Cistron | Exon |
| Cistron is segments of DNA that possess both exon and intron. | Exons are coding regions of a gene that code for different proteins. |
| It refers to the whole gene. | It refers to a part of gene. |
Question 31.
Differentiate between exons and introns. (All India 2012C)
Answer:
Differences between exons and introns are as follows
| Exons | Introns |
| Regions of a gene which become part of mRNA. | Regions of a gene which do not form part of mRNA. |
| Code for the different proteins and hence called coding sequence. | Removed during the processing of mRNA because they are non-coding sequences. |
Question 32.
State the dual role of deoxyribonucleoside triphosphates during DNA replication. (Delhi 2011)
Answer:
- The deoxyribonucleoside triphosphates are the building blocks for the DNA strand (polynucleotide chain), i.e. they act as substrates.
- These also serve as energy source in the form of ATP and GTP because of the presence of high energy terminal phosphate groups.
Question 33.
Answer the questions based on the dinucleotide shown below.
(i) Name the type of sugar guanine base is attached to.
(ii) Name the linkage connecting the two nucleotides.
(iii) Identify the 3′ end of the dinucleotide. Give a reason for your answer. (All India 2010C)
Answer:
(i) Pentose sugar or deoxyribose sugar.
(ii) Two nucleotides are linked through 3′-5′ phosphodiester linkage to form a dinucleotide.
(iii) The ribose sugar has a free 3′ – OH group which is referred to as 3’end of the polynucleotide chain.
Question 34.
Make a labelled diagram of an RNA dinucleotide showing its 3′ → 5′ polarity. (All India 2010C)
Answer:
RNA dinucleotide.
Question 35.
Answer the following questions based on Meselson and Stahl’s experiment on E.coli.
(i) Write the name of the chemical substance used as the only source of nitrogen in the experiment.
(ii) Why did they allow the synthesis of the light and the heavy DNA molecules in the organism?
(iii) How did they distinguish the heavy DNA molecules from the light DNA molecules?
(iv) Write the conclusion the scientists arrived at, at the end of the experiment. (All India 2019,11)
Answer:
(i) NH4Cl (Ammonium chloride) with N14 atom.
(ii) It is to show that after one generation E. coli with 15N -DNA in a medium of 14N, has DNA of intermediate density between the light and heavy DNAs.
It shows that of the two strands, only one strand is synthesised newly, using the 14N-nitrogen source in the medium.
(iii) The heavy and light DNA molecules can be differentiated by centrifugation in a cesium chloride (CsCl) density gradient.
The 15N -DNA was heavier than 14N -DNA and the hybrid 15N – 14N -DNA was intermediate between the two newly form DNA strands.
(iv) Scientists concluded that the DNA replication is semi-conservative, i.e. of the two strands of DNA, one is the parental strand, while the other is newly synthesised.
Question 36.
Explain the mechanism of DNA replication with the help of a replication fork. What role does the enzyme DNA-ligase play in a replication fork? (Delhi 2019)
Answer:
Replication in DNA strand occurs within a small opening of the DNA helix, known as replication fork.
For figure, see Answer No. 26.
DNA polymerases catalyse polymerisation only in one direction, i.e. 5′ → 3′. It creates additional complications at the replicating fork. Consequently, on one strand (template 3′ → 5′), the replication is continuous. This is known as leading strand, while on the other strand (template 5′ → 3′), it is discontinuous. This is known as lagging strand.
The discontinuously synthesised fragments called Okazaki fragments are later joined by DNA ligase.
Question 37.
Construct and label a transcription unit from which the RNA segment given below has been transcribed. Write the complete name of the enzyme that transcribed this RNA. (Delhi 2019)
Answer:
The process of copying genetic information from one strand of DNA into RNA is called ‘transcription’. Transcription is catalysed by ‘DNA dependent RNA polymerase’.
The RNA molecule given in question should be
As RNA have uracil in place of thymine.
For given RNA, the transcription unit will be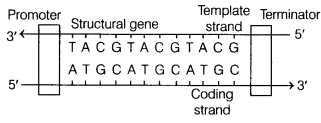
Question 38.
Why is DNA molecule considered as a better hereditary material than RNA molecule? (2018C)
Or
Why is DNA a bettter genetic material when compared to RNA? (Delhi 2015C)
Or
Why is DNA considered a better hereditary material than RNA? (Foreign 2012)
Answer:
DNA is considered a better genetic material when compared to RNA because of its high stability and low reactivity. RNA being more reactive, is labile and easily degradable.
The high reactivity of RNA is contributed by the 2-OH group in the nucleotides. RNA as a catalyst (e.g. ribozyme) is also more reactive. Uracil present in RNA makes it unstable over DNA that contains thymine.
Question 39.
Name the three polymerases found in eukaryotic’cells and mention their functions. (2018C)
Answer:
Three types of RNA polymerases are found in eukaryotic cells and their functions are as follows
- RNA polymerase-I transcribes rRNAs.
- RNA polymerase-II transcribes the precursor of mRNA called hnRNA.
- RNA polymerase-III transcribes tRNA, 5 SrRNA and swRNAs.
Question 40.
Explain the post-transcriptional modifications the AnRNA undergoes in eukaryotic. (2018C)
Answer:
Refer to page no. 138-139.
Question 41.
Study the diagram given below:
Name the linkage X, Y, Z and the respective molecules formed by them. (Delhi 2016C)
Answer:
X : N-glycosidic linkage. It forms nucleoside.
Y : Phosphoester linkage. It forms nucleotide.
Z : 3′-5’phosphodiester linkage. It forms polynucleotide.
Question 42.
Describe the experiment that helped demonstrate the semi-conservative mode of DNA replication. (All India 2016)
Or
How was a heavy isotope of nitrogen used to provide experimental evidence to semi-conservative mode of DNA replication? (Foreign 2015)
Answer:
Matthew Meselson and Franklin Stahl conducted an experiment with Escherichia coli (1958) as follows
- They grew many generations of E. coli in a medium that contained 15NH4Cl (15N is the heavy isotope of nitrogen) as the only source of nitrogen. The result was that 15N was incorporated into the newly synthesised DNA. Upon centrifugation in a cesium chloride (CsCl) density gradient, this heavy DNA molecule could be distinguished from the normal DNA.
- The cells were then transferred into a medium containing normal 14NH4Cl.
- At definite time intervals, as the cells multiplied, samples were taken and the DNA which remained as double-stranded helices were extracted.
- The samples were separated independently on CsCl gradient to measure the densities of DNA.
- The DNA obtained from the culture, one generation after the transfer from 15N to 14N medium had a hybrid or intermediate density.
- The DNA obtained from the culture after another generation (generation II), was composed of equal amounts of hybrid DNA and Tight’ DNA.
Thus, Meselson and Stahl concluded that the DNA replication is semi-conservative, i.e. out of the two strands of DNA one is the parental
strand, while another is newly synthesised.
Question 43.
(i) Differentiate between a template strand and coding strand of DNA.
(ii) Name the source of energy for the replication of DNA. (Delhi 2015C)
Answer:
(i) Differences between template strand and coding strand are as follows
| Template strand | Coding strand |
| It has 3′- 5’polarity. | It has 5′- 3’polarity. |
| It gets transcribed. | It does not get transcribed. |
| Its sequence is complementary to mRNA formed. | Its sequence is same as mRNA except it contains ‘T’ instead of ‘U’. |
(ii) The source of energy for the replication of DNA are the deoxyribonucleoside triphosphates that have two terminal high energy phosphates.
Question 44.
(i) A DNA segment has a total of 1000 nucleotides, out of which 240 of them are adenine containing nucleotides. How many pyrimidine bases this DNA segment possesses?
(ii) Draw a diagrammatic sketch of a portion of DNA segment to support your answer. (Delhi 2015)
Or
(i) A DNA segment has a total of 1500 nucleotides, out of which 410 are guanine containing nucleotides. How many pyrimidine bases this segment possesses?
(ii) Draw a diagrammatic sketch of a portion of DNA segment to support your answer. (All India 2015)
Or
(i) A DNA segment has a total of 2000 nucleotides, out of which 520 are adenine containing nucleotides. How many purine bases this DNA segment possesses?
(ii) Draw a diagrammatic sketch of a portion of DNA segment to support your answer. (Delhi 2014)
Answer:
(i) According to Chargaff’s rule, ratio of purines to pyrimidines is equal,
i.e. A + G = C + T
The number of adenine (A) is equal to the number of thymine (T).
A = 240(given)
Therefore, T = 240
Also, the number of guanine (G) is equal to cytosine (C).
Thus, G + C = 1000 – [A + T]
G + C = 1000 – 480= 520
Hence, G = 260, C = 260
The number of pyrimidine bases,
i.e. C + T= 240 + 260 = 500
(ii) Diagrammatic sketch of portion of DNA segment
Or
(i) Given, G = 410 therefore, C = 410
A + T = 1500 – (G + C)
= 1500 – 820 = 680
Hence, A = 340; T = 340
The number of pyrimidine bases, i.e.
C + T = 410 + 340 = 750
(ii) Refer to above figure.
Or
(i) Given, A = 520
therefore, T = 520
A + T = 520 + 520 = 1040
Total number of nucleotides = 2000
G + C = 2000 – 1040 = 960
G = 960/2 = 450
Hence, total number of purine bases are
⇒ A + G = 520 + 480 = 1000
(ii) Refer to above figure.
Question 45.
With the help of a schematic diagram, explain the location and role of the following in a transcription unit. Promoter, structural gene, terminator. (All India 2014C)
Answer:
Structure of a transcription unit
The promoter and terminator flank the structural gene in a transcription unit. The promoter is located towards 5’end (upstream) of the structural gene and it helps to initiate transcription by binding with RNA polymerase. The terminator is located toward 3’end (downstream) of the coding strand and it usually defines the end of the process of transcription. The structural gene is present in between promoter and terminator. It codes for enzyme or protein for structural functions.
Question 46.
(i) What are the transcriptional products of RNA polymerase-III?
(ii) Differentiate between capping and tailing.
(iii) Expand ArcRNA. (All India 2014C)
Answer:
(i) RNA polymerase-III is responsible for the transcription of tRNA, SrRNA and snRNAs (small nuclear RNAs).
(ii) In capping process, an unusual nucleotide (methyl guanosine triphosphate) is added to ‘5’ end of hnRNA.
In tailing process, 200-300 adenylate residues are added at 3′ end of hnRNA.
(iii) hnRNA is heterogeneous nuclear RNA.
Question 47.
It is established that RNA is the first genetic material. Explain giving three reasons. (Delhi 2012)
Answer:
RNA is the first genetic material in cells because
- RNA is capable of both storing genetic information and catalysing chemical reactions.
- Essential life processes (such as metabolism, translation, splicing, etc.) were evolved around RNA.
- It shows the power of self-replication.
Question 48.
(i) Construct a complete transcription unit with promoter and terminator on the basis of hypothetical template strand given below.
(ii) Write the RNA strand transcribed from the above transcription unit along with its polarity. (Delhi 2012)
Answer:
(i) Transcription unit
(ii) RNA strand transcribed from the above transcriptional unit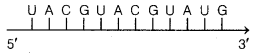
Question 49.
List the salient features of double helix structure of DNA. (All India 2012)
Answer:
Salient features of DNA double helix are as follows
- It is made up of two polynucleotide chains containing the backbone of sugar-phosphate from which the bases project inside.
- The two chains have anti-parallel polarity, i.e. one of them is 5′ → 3′, the other has 3′ → 5’polarity.
- The bases in two strands are paired through hydrogen bond (H-bonds) forming base pairs (bp). Adenine pairs through two hydrogen bonds with thymine on opposite strand and vice-versa. In the same way, guanine is bonded with cytosine through three H-bonds. Due to which, purine always comes opposite to a pyrimidine.
- The two chains are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm and there are roughly 10 bp in each turn. Consequently, the distance between base pair in a helix is about 0.34 nm.
- The plane of one base pair stacks over the other in double helix. This confers stability to the helical structure.
Question 50.
How is hnRNA processed to form mRNA? (Foreign 2012)
Answer:
The precursor of mRNA transcribed by RNA polymerase-II is called heterogeneous nuclear RNA (hnRNA). It undergoes following processing to form nascent wRNA.
- Splicing In this process, the non-coding introns are removed and coding sequences called exons are joined in a definite order. This is required because primary transcript contains both introns and exons.
- Capping Refer to Answer No. 46.
- Tailing Refer to Answer No. 46.
- The fully processed mRNA is released from the nucleus into cytoplasm for translation.
Question 51.
The base sequence in one of the strands of DNA is TAGCATGAT.
(i) Give the base sequence of the complementary strand.
(ii) How are these base pairs held together in a DNA molecule?
(iii) Explain the base complementarity rule. Name the scientist who framed this rule. (Delhi 2011)
Answer:
(i) ATCGTACTA.
(ii) Base pairs are held together by weak hydrogen bonds, adenine pairs with thymine by two H-bonds and guanine pairs with cytosine through three H-bonds.
(iii) Base complementarity rule : For a double-stranded DNA, this rule states that the ratio between adenine and thymine and that between guanine and cytosine are constant and equal to one. Erwin Chargaff framed this rule.
Question 52.
Why do you see two different types of replicating Strands in the given DNA replication fork? Explain. Name these strands. (Delhi 2011)
Answer:
Two different types of parent strands function as template strands.
On the template strand with 3′ → 5’polarity, the new strand is synthesised as a continuous strand because the enzyme DNA polymerase can carry out polymerisation of the nucleotides only in 5′ → 3′ direction. This is called continuous synthesis and the strand is called leading strand.
On the template strand with 5′-» 3’polarity, the new strand is synthesised from the point of replication fork in short stretches called Okazaki fragments and this strand is called lagging strand. Okazaki fragments are later joined by DNA ligases to form a continuous strand.
Question 53.
Monocistronic structural genes in eukaryotes have interrupted coding sequences. Explain. How are they different in prokaryotes? (Delhi 2011C)
Answer:
Monocistronic structural genes in eukaryotes have interrupted coding sequences due to the presence of introns, i.e. non-coding sequences.
Differences between monocistronic structural gene in prokaryotes and in eukaryotes are as follows
| Structural gene in prokaryotes | Structural gene in eukaryotes |
| Consists of only coding sequences. | Consists of both coding and non-coding sequences. |
| Information is continuous as only exons are present. | Information is split due to the presence of introns in between exons. |
| There is no need of splicing. | Splicing is required to make functional mRNA. |
Question 54.
Explain the role of 35S and 32P in the experiments conducted by Hershey and Chase. (All India 2010C)
Answer:
Hershey and Chase used 35S and 32P in their culture medium. These are radioactive sulphur and phosphorus, respectively. These two components were used to detect whether the genetic material is DNA or protein.
Role of 32P and 35S Viruses grown on medium with 32S. had non-radioactive genetic material and radioactive protein as sulphur is not present in DNA but found in protein.
While those grown on 32P had radioactive genetic material because DNA contains phosphorus but proteins does not contain it. Thus, it was established that DNA is the genetic material.
Question 55.
Describe the initiation process of transcription in bacteria. (Delhi 2010)
Answer:
Initiation process of transcription in bacteria RNA polymerase becomes associated transiently to an initiation factor sigma (cr) and binds to specific sequence on DNA called promoter to initiate transcription.
A single DNA-dependent RNA polymerase catalyses the transcription of all the three types of RNA.
Question 56.
Describe the elongation process of transcription in bacteria. (Delhi 2010)
Answer:
Elongation process of transcription in bacteria RNA polymerase facilitates opening of the DNA helix after binding to promoter. It uses ribonucleoside triphosphates as substrate and polymerises the nucleotides in a template dependent fashion following complementarity. The process continues till RNA polymerase reaches the terminator region on the DNA strand.
Question 57.
Describe the termination process of transcription in bacteria. (Delhi 2010)
Answer:
Termination occurs when RNA polymerase reaches the terminator region and the nascent RNA falls off. The RNA polymerase being transiently associated with termination factor rho (p) also falls off the transcription unit.
Question 58.
Draw a schematic representation of a dinucleotide. Label the following :
(i) The component of a nucleotide
(ii) 5′ end
(iii) N-glycosidic linkage
(iv) Phosphodiester linkage (Foreign 2010)
Answer:
Schematic representation of a dinucleotide.
Question 59.
Describe the role of RNA polymerases in transcription in bacteria and in eukaryotes. (Foreign 2010)
Answer:
In bacteria, there is a single DNA-dependent RNA polymerase, which catalyses the formation of mRNA, tRNA and rRNA.
In eukaryotes, there are three types of RNA polymerases, which show division of labour.
These are as follows
- RNA polymerase-I transcribes rRNAs, (28S, 18S and 5.8S)
- RNA polymerase-II transcribes the precursor of mRNA called hnRNA.
- RNA polymerase-III transcribes tRNA, 5 SrRNA and snRNAS.
Question 60.
(i) State the ‘central dogma’ as proposed by Francis Crick.
(ii) Explain, how the biochemical characterisation (nature) of ‘Transforming Principle’ was determined, which was not defined from Griffith’s experiments. (2018)
Answer:
(i) Central Dogma It states that there is one way or unidirectional flow of information from master copy DNA to working copy RNA (transcription) and from working copy RNA to building plan of polypeptide chain (translation).
Central dogma of molecular biology was proposed by Crick (1958). It can also be written as follows![]()
Exception Reverse transcription is an exception to central dogma. The discovery of ‘RNA dependent DNA polymerase’ gave the concept of central dogma reverse or teminism. It suggested that information flow can also take place from RNA to DNA.
During this, transcription or RNA synthesis occurs over DNA and translation or protein synthesis occurs over ribosomes. In eukaryotes, these two processes are separated in time and space. This protects DNA from respiratory enzymes and RNA’s degradation from nucleases.
(ii) Various experiments were conducted to prove that DNA is the genetic material. Griffith did his experiment and concluded that some substance causes transformation. However, he did not tell the substance that had caused the transformation.
Biochemical Characterisation of Transforming Principle Three scientists Avery, MacLeod and McCarty revealed that the chemical nature of the transforming substance was DNA. They showed that DNA isolated from S-strain could itself confer the pathogenic properties to R-strain.
This fact suggested that DNA possesses the genetic properties.
The outline of the experiment is given below
Thus, on the basis of their experiment, they concluded that DNA is the hereditary material.
Question 61.
(i) Why does DNA replication occur in small replication forks and not in its entire length?
(ii) Why is DNA replication continuous and discontinuous in a replication fork?
(iii) State the importance of origin of replication in a replication fork. (2018C)
Answer:
(i) DNA replication occurs in small replication fork and not in its entire length because whole DNA cannot be opened in one stretch due to high energy requirement.
(ii) DNA replication is continuous and discontinuous in a replication fork because the enzyme DNA polymerase can carry out polymerisation of the nucleotides only in 5-3′ direction. On the template strand with 3-5′ polarity, DNA replication is continuous. On the template strand with polarity 5′-3′, the DNA replication occurs in short stretches and is called discontinuous.
(iii) Replication of DNA does not initiate randomly, and DNA polymerases on their own cannot initiate replication. So, there is a need of specific sequence, called origin of replication from which the replication starts. DNA polymerase binds to it and continues the process.
Question 62.
Answer the following questions based on Hershey and Chase’s experiments
(i) Name the kind of virus they worked with and why?
(ii) Why did they use two types of culture media to grow viruses in? Explain.
(iii) What was the need for using a blender and later a centrifuge during their experiments?
(iv) State the conclusion drawn by them after the experiments. (All India 2016)
Or
How did Hershey and Chase established that DNA is transferred from virus to bacteria? (Delhi 2015)
Or
Describe the Hershey and Chase’s experiment. Write the conclusion drawn by the scientists after their experiment. (All India 2014)
Or
Name the scientists, who proved experimentally that DNA is the genetic material. Describe their experiment. (Delhi 2014)
Or
(i) Describe Hershey and Chase’s experiment.
(ii) Write the aim of the experiment. (Delhi 2010C; All India 2010)
Answer:
(i) They worked with bacteriophage, i.e. viruses that infect bacteria. These viruses were used because during infection they transfer their genetic material into bacteria.
(ii) They used two types of culture media, containing 35S and 32P, so as to compare that which one out of DNA and proteins gets transferred from virus to bacteria and acts as genetic material.
(iii) A blender and centrifuge was used to open up the bacterial cells and viral particles, so, that genetic material could be exposed.
(iv) Conclusion DNA is the genetic material.
Or
Alfred Hershey and Martha Chase (1952) established that DNA is transferred from viruses that infect bacteria. In their experiment, bacteriophages (viruses that infect bacteria) were used. They grew some viruses on a medium that contained radioactive phosphorus and some others on medium containing radioactive sulphur. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA, but not radioactive protein because DNA contains phosphorus, but protein does not. In the same way, viruses grown on radioactive sulphur contained radioactive protein, but not radioactive DNA because DNA does not contain sulphur. Radioactive phages were allowed to attach to E.coli bacteria. As the infection proceeded, viral coats were removed from the bacteria by agitating them in a blender. The virus particles were seperated from the bacteria by spinning them in centrifuge.
Bacteria which were infected with viruses that had radioactive DNA were radioactive, indicating that DNA was the material that passed from the virus to the bacteria. Bacteria that were infected with viruses that had radioactive proteins were not radioactive. This indicated that the proteins did not enter the bacteria from viruses. It proved that DNA is the genetic material that is passed from virus to bacteria.
Question 63.
(i) Name the stage in the cell cycle where DNA replication occurs.
(ii) Explain the mechanism of DNA replication. Highlight the role of enzymes in the process.
(iii) Why is DNA replication said to be semi-conservative? (All India 2016)
Or
(i) Explain the process of DNA replication with the help of a schematic diagram.
(ii) In which phase of the cell cycle does replication occur in eukaryotes? What would happen if cell division is not followed after DNA replication? (Delhi 2014)
Or
(i) Draw a labelled diagram of a ‘replicating fork’ showing the polarity. Why does DNA replication occur within such ‘forks’?
(ii) Name two enzymes involved in the process of DNA replication, along with their properties. (Outside Delhi 2015)
Answer:
(i) During the synthetic phase of interphase of cell cycle DNA duplicates or replicates.
(ii) Process of DNA Replication Replication is an energy expensive process, deoxyribonucleoside triphosphate serves the dual purpose of
- acting as a substrate.
- providing energy (from two terminal phosphates).
In a long DNA molecule, replication takes place within a small opening of the DNA helix, known as replication fork because whole DNA does not open in one stretch due to high energy requirement. DNA dependent DNA polymerases catalyse polymerisation only in one direction, i.e. 5′ → 3′. This brings additional complications at the replication fork.
On one strand (template with polarity 3′ → 5′), replication is continuous, while on the 5′ → 3′ strand, replication is discontinuous.
Thus, the fragments synthesised are discontinuous and later joined by the enzyme DNA ligase.
The DNA polymerases cannot initiate replication process on their own and replication does not initiate randomly anywhere in DNA.
It begins at definite regions in a DNA molecule known as origin of replication (Ori).
Enzymes for DNA Replication
The process of replication requires a set of catalysts (enzymes) as given below
(a) DNA-Dependent DNA Polymerase: It is the main enzyme which uses a DNA template to catalyse the polymerisation of deoxynucleotides. The average rate of polymerisation is 2000 bp (base pairs) per second approximately. These enzymes are highly efficient as they have to catalyse polymerisation of a large number of nucleotides in a very short time. These polymerases act very fast, catalyse the replication process with high degree of accuracy as any mistake would result in mutations.
(b) Helicase It unwinds the DNA strand, i.e. separates the two strands from one point for the formation of a replication fork.
(c) DNA Ligase It facilitates the joining of DNA strands together by catalysing the formation of phosphodiester bond. It plays a role in repairing single strand breaks in duplex DNA.
(iii) It is said to be semi-conservative because in newly synthesised DNA, one strand is parental and one is new, so it conserves the one strand. DNA replication is followed by cell division. In case, the latter fails to occur, polyploidy may occur.
Question 64.
List the criteria of a molecule that can act as genetic material must fulfil. Which one of the criteria is best fulfilled by DNA or by RNA thus making one of them a better genetic material than the other? Explain. (All India 2016)
Answer:
From the Hershey and Chase experiment, the fact was established that DNA acts as genetic material. But later studies revealed that in some viruses (e.g. Tobacco Mosaic Viruses, QB bacteriophage, etc.) RNA is the genetic material. Following are the criteria that a molecule must fulfil to act as a genetic material.
- It- should be able to replicate.
- It should be chemically and structurally stable.
- It should provide the scope for slow changes (mutation), which are required for evolution.
- It should be able to express itself in the form of ‘Mendelian characters’.
According to these criteria, both DNA and RNA have the ability to direct their duplications (because of the rule of base pairing and complementarity).
So, both the nucleic acids (DNA and RNA) have the ability to direct their duplications, whereas the other molecules in the living system, fail to fulfil first criteria itself, e.g. protein. The most important criteria of genetic material is the stability as the genetic material should not change with the different stages of life cycle, age or with change in the physiology of an organism. Both DNA and RNA have the ability to mutate. Since, RNA is unstable, it mutates at a faster rate. That is why, those viruses, which have RNA genome and a shorter lifespan, undergo mutation and thus, evolve rapidly.
DNA is dependant on RNA for protein synthesis, whereas RNA directly codes for protein synthesis. This proves that both RNA and DNA act as genetic material, but DNA being more stable is preferred for the storage of genetic information.
Question 65.
Describe Meselson and Stahl’s experiment that was carried in 1958 on E. coli. Write the conclusion they arrived at after the experiment. (All India 2016)
Or
(i) What is central dogma? Who proposed it?
(ii) Describe Meselson and Stahl’s experiment to prove that the DNA replication is semi-conservative. (Outside Delhi 2015C)
Or
‘DNA replication is semi-conservative’. Name the scientists who proposed it and who proved it. How was it proved experimentally? (All India 2014C)
Or
Describe Meselson and Stahl’s experiment and write the conclusion they arrived at. (Foreign 2014: Delhi 2012)
Answer:
Refer to Answer No. 42.
Or
Central Dogma Francis Crick proposed the central dogma in molecular biology, which states that the genetic information flows from DNA → RNA → Proteins.
Question 66.
(i) How are the following formed and involved in DNA packaging in the nucleus of a cell?
(a) Histone octamer
(b) Nucleosome
(c) Chromatin
(ii) Differentiate between euchromatin and heterochromatin. (All India 2016)
Answer:
(i) (a) Histone Octamer Histones are the proteins that are rich in basic amino acids, i.e. lysine and arginine. Both these amino acids carry positive charges in their side chains. So, histones are a set of positively charged, basic proteins (histones + protamine). A histone octamer is formed by the organisation of two molecules each of H2A, H2B, H3 and H4 histones so asa to make a unit of 8 molecules. It helps to package DNA into nucleosome.
(b) Nucleosome The negatively charged DNA is wrapped around the positively charged histone octamer, forming a structure known as nucleosome. It forms the fundamental repeating units of eukaryotic chromatin. It is used to pack the large eukaryotic genome into the nucleus.
(c) Chromatin One nucleosome contains 200 base pairs of DNA helix, approximately. Nucleosomes are the repeating unit of chromatin, which are thread-like stained (coloured) bodies present in nucleus.
The nucleosomes in chromatin look like ‘beads-on-string’ when observed under an electron microscope. The chromatin is further packed to form a solenoid structure of 30 mm diameter and further supercoiling tends to form a looped structure called chromatin fibre and then chromatid. This further coils and condenses at metaphase stage to form the chromosomes.
(ii) Differences between euchromatin and heterochromatin are as follows
| Euchromatin | Heterochromatin |
| It is lightly stained region. | It is darkly stained region of the chromatin (chromosome). |
| It is loosely coiled region and thus, has less DNA. | It is the compactly coiled region and thus, has more DNA. |
| It is transcriptionally active and is transcribed into mRNA. | It is transcriptionally inert and cannot be transcribed into mRNA due to very tight coiling. |
Question 67.
(i) Explain with the help of Griffith’s experiment how the search for genetic material was conducted and what was the conclusion drawn?
(ii) How did MacLeod, McCarty and Avery establish the biochemical nature of the so called ‘genetic material’ identified by Griffith in his experiment? (Delhi 2016C)
Or
(i) Describe the series of experiments of F Griffith. Comment on the significance of the results obtained, (ii) State the contribution of MacLeod, McCarty and Avery. (All India 2016, 2010)
Or
(i) Describe the experiment which demonstrated the existence of transforming principle.
(ii) How was the biochemical nature of this transforming principle determined by Avery, MacLeod and McCarty? (Foreign 2015)
Or
(i) Describe the various steps of Griffith’s experiment that led to the conclusion of the ‘transforming principle’.
(ii) How did the chemical nature of the ‘transforming principle’ get established? (All India 2014)
Or
(i) Write the conclusion drawn by Griffith at the end of his experiment with Streptococcus pneumoniae.
(ii) How did O Avery, C MacLeod and M McCarty prove that DNA was the genetic material? Explain. (All India 2013)
Or
Describe Frederick Griffith’s experiment on Streptococcus pneumoniae. Discuss the conclusion he arrived at. (All India 2012)
Answer:
(i) Griffith Experiments: It took a long time to discover and prove that DNA acts as a genetic material. By 1926, the quest to determine the mechanism for genetic inheritance had reached the molecular level. Transforming Principle Frederick Griffith in 1928, carried out a series of experiments with Streptococcus pneumoniae (a bacterium that causes pneumonia).
He observed that when these bacteria (Streptococcus pneumoniae) were grown on a culture plate, some of them produced smooth, shiny colonies (S-type), whereas the others produced rough colonies (R-type).
This difference in appearance of colonies (smooth/rough) was due to the presence or absence of mucus (polysaccharide) coat.
In his experiments, he first infected two separate groups of mice as follows S-strain (virulent strain) → Inject into mice → Mice die.
R-strain (non-virulent strain) → Inject into mice → Mice live.
S-strain (heat-killed) → Inject into mice → Mice live.
S-strain (heat-killed) + R-strain (live) → Inject into mice → Mice die.
Griffith concluded that the live R-strain, bacteria, had been transformed by the heat-killed S-strain bacteria. He concluded that, there was some ‘transforming agent that was transferred from the heat-killed S-strain, which helped the R-strain bacteria to synthesise a smooth polysaccharide coat and thus, become virulent.
He concluded that must be due to the transfer of the genetic material. However, he was not able to define the biochemical nature of genetic material from his experiments.
(ii) Contributions of MacLeod, McCarty and Avery : These scientists worked to determine the chemical nature of transforming principle of Griffith’s experiments. They discovered that protein digesting enzymes (proteases) and RNA-digesting.enzymes (RNase) did not affect transformation. Digestion with DNase did inhibit transformation suggesting that DNA was responsible for the transformation.
Question 68.
(i) Describe the process of trancription in bacteria.
(ii) Explain the processing the /mRNA needs to undergo before becoming functional mRNA in eukaryotes. (Outside Delhi 2016)
Or
Describe the process of transcription in bacterium. (All India 2014C)
Answer:
(i) The process of transcription in prokaryotes consists of three steps
- Initiation Refer to Answer No. 55.
- Elongation Refer to Answer No. 56.
- Termination Refer to Answer No. 57.
(ii) In eukaryotes, the primary transcript is often larger than the functional RNA, called heterogeneous nuclear RNA or hnRNA.
Post-transcription processing is necessary to convert primary transcript of different types of RNAs into functional RNAs.
The processing includes
- Cleavage Larger RNA precursors are cleaved to form smaller RNAs.
- Splicing Eukaryotic transcripts possess extra segments called introns which are non-functional. They do not appear in mature or processed RNA. The functional coding sequences are called exons. Splicing is the removal of introns and fusion of exons in a definite order to form functional RNAs.
- Terminal additions (capping and tailing) Capping includes addition of an unusual nucleotide to 5’ end of hrRNA. These unusual segments are CCA segment in tRNA and cap nucleotides at 5’end of mRNA. In tailing, poly-A segment (200-300 residues) are added at 3’end of mRNA. Cap is formed by the modification of GTP into 7-methyl guanosine or 7 mG.
- Nucleotide modifications They are most common in tRNA-methylation (i.e. methyl cytosine, methyl guanosine), deamination (e.g. inosine form adenine), dihydrouracil, pseudouracil, etc.
Question 69.
Explain the process of transcription in prokaryotes. How is the process different in eukaryotes? (Outside Delhi 2015)
Answer:
In prokaryotes or bacterial cell, transcription occurs in three steps-initiation elongation and termination. For detail, Refer to text on pages 138-139.
Question 70.
(i) Explain the role of DNA-dependent RNA polymerase in initiation, elongation and termination during transcription in bacterial cell.
(ii) How is transcription a more complex process in eukaryotic cells? Explain. (Foreign 2011)
Answer:
(i) Role of DNA-dependent RNA polymerase
- RNA polymerase becomes associated transiently with initiation factor (σ) and binds to the promoter site on DNA and initiates transcription.
- It uses the nucleoside triphosphate as substrates and polymerises them in a template-dependent fashion following the base complementarity rule in the 5′ → 3′ direction.
- It also facilitates the opening of the DNA helix and continues the elongation process.
- When the polymerase falls off a terminator region on the DNA, the nascent RNA separates. This results in termination.
(ii) Reasons that transcription is more complex in eukaryotes are
(a) The three types of RNA polymerases in the nucleus show division of labour.
- RNA polymerase-I transcribes rRNAs (28S, 18S and 5.8S).
- RNA polymerase-II transcribes the precursor of mRNA, called hnRNA.
- RNA polymerase-III transcribes tRNA, 5 SrRNA and snRNAse.
(b) hnRNA contains both coding sequences called exons and non-coding sequences called introns. So, it undergoes a process called splicing, in which the non-coding sequences (introns) are removed and the coding sequences (exons) are joined together in a defined order.
(c) In capping, unusual nucleotide, methyl guanosine triphosphate residues are added at the 5’end of the hnRNA.
(d) In tailing, 200-300 adenylate residues are added at the 3’ end of the hnRNA.
Question 71.
(i) Mention the contributions of the following scientists:
(a) Maurice Wilkins and Rosalind Franklin
(b) Erwin Chargaff
(ii) Draw a double-stranded dinucleotide chain with all the four nitrogen bases. Label the polarity and the components of the dinucleotide. (All India 2011C)
Answer:
(i) (a) Maurice Wilkins and Rosalind: Franklin They carried out X-ray diffraction studies on the structure of DNA molecule.
(b) Erwin Chargaff He proposed the base complementarity rule, according to which
(ii) Refer to Answer No. 28.
Question 72.
Study the flowchart given below and answer the questions that follows
I. S-strain → into mice → mice die
II. R-strain → into mice → mice live
III. Heat-killed S-strain + Live R-strain → into mice → A
IV. Heat-killed S-strain + DNase + Live R-strain → into mice → B
(i) Name the organism and differentiate between its two strains S and R, respectively.
(ii) Write the result A and B obtained in step III and IV, respectively.
(iii) Name the scientist who performed the.steps I, II and III.
(iv) Write the specific conclusion drawn from the step IV. (All India 2010C)
Answer:
(i) The organism is bacterium Streptococcus pneumoniae.
Differences between S-strain and R-strain are as follows
| S-strain | R-strain |
| They form smooth colonies protected by a capsule. | They form rough colonies without a capsule. |
| They are virulent. | They are non-virulent. |
(ii) 1 A – Mice died.
B – Mice lived.
(iii) Frederick Griffith performed these steps.
(iv) When DNase was added to the medium, the DNA of the heat-killed cells got denatured and was unable to carry transformation. This indicates that DNA was the transforming component.
Question 73.
Name one amino acid, which is coded by only one codon. (2018C)
Answer:
Methionine.
Question 74.
Give an example of a codon having dual function. (All India 2016)
Answer:
AUG is a codon with dual functions. It codes for the amino acid methionine (met) and also acts as an initiator codon of polypeptide synthesis during protein synthesis.
Question 75.
Write the two specific codons that a translational unit of mRNA is flanked by one on either sides. (Outside Delhi 2015C)
Answer:
Two specific-codons that are flanked on either sides of mRNA in a translation unit are
- Start codon (AUG)
- Stop codon (UAG or UAA or UAA).
Question 76.
How does a degenerate code differ from an unambiguous one? (Foreign 2015)
Answer:
When some amino acids are coded by more than one codon, the code is said to be degenerate.
On the other hand, when one codon codes for only one amino acid, it is called unambiguous.
Question 77.
How is repetitive/satellite DNA separated from bulk genomic DNA for various genetic experiments? (Delhi 2014)
Answer:
Satellite DNA is separated from bulk genomic DNA by density-gradient centrifugation technique.
Question 78.
State which human chromosome has
(i) the maximum number of genes and
(ii) the one which has the least number of genes. (Foreign 2014,2011)
Answer:
(i) Chromosome 1 (2968 genes).
(ii) Chromosome Y (231 genes).
Question 79.
Which one of the two subunits of ribosome encounters an mRNA? (Delhi 2013c)
Answer:
Smaller subunit.
Question 80.
Differentiate between the following: Inducer and repressor in lac operon. (Delhi 2011C)
Answer:
Inducer It is a chemical which after coming in contact with repressor, changes it into non-DNA binding state, so as to free the operator gene.
Repressor It is a regulator protein meant for blocking the operator gene.
Question 81.
Mention the role of the codons AUG and UGA during protein synthesis. (Delhi 2011,10)
Answer:
AUG acts as initiation codon and codes for amino acid methionine.
UGA acts as stop/termination codon that signals termination of polypeptide synthesis.
Question 82.
Mention the contribution of genetic maps in human genome project. (All India 2011)
Answer:
Genetic maps are used as a starting point in the sequencing of whole genome.
![]()
Question 83.
Mention any two ways in which Single Nucleotide Polymorphisms (SNPs) identified in human genome, can bring out revolutionary changes in biological and medical sciences. (All India 2011c)
Answer:
- By tracing human history.
- By finding chromosomal locations for disease associated sequences.
Question 84.
Differentiate between the genetic codes given below
(i) Unambiguous and universal
(ii) Degenerate and initiator (All india 2017)
Answer:
(i) The difference between unambiguous and universal genetic codes is as follows
Unambiguous:
In genetic code, one codon codes for only one amino acid, hence it is unambiguous.
Universal:
The genetic code is universal, i.e. each codon codes for same amino acid in all organisms.
(ii) The difference between degenerate and initiator codes is as follows
| Degenerate | Initiator |
| Some amino acids are coded by more than one codon, hence the code is degenerate. | These codons act as start signal for translation, |
| e.g serine, leucine, arginine are encoded by 6 codons. | e.g. AUG acts as initiator codon and it codes for methionine. |
Question 85.
Following are the features of genetic codes. What does each one indicate? (All India 2016)
Stop codon, Unambiguous codon, Degenerate codon, Universal codon.
Answer:
- Stop codon does not code for any amino acids, e.g. UGA.
- Unambiguous codon codes for only one . amino acid, e.g. CCG codes for proline.
- Degenerate codon Genetic code is described as degenerate when a single amino acid is coded by more than one codon, e.g. serine is coded by 6 codons.
- Universal codons codes for the same amino acid in all organisms (except in mitochondria and few protozoan).
Question 86.
What is aminoacylation? State its significance. (All India 2016)
Or
Explain aminoacylation of tRNA. (All India 2014C)
Answer:
Aminoacylation or charging of tRNA is a process in which amino acids get activated in the presence of ATP and get linked to their cognate tRNA.
Significance of aminoacylation The charged tRNA carries amino acids to the site of protein synthesis. If two charged tRNAs are brought close to each other, the formation of peptide bond is favoured energetically.
Question 87.
State the functions of ribozyme and release factor in protein synthesis, respectively. (Outside Delhi 2015C)
Answer:
Ribozyme in bacteria is 23S rRNA, that acts as an enzyme for the formation of a peptide bond between two amino acids.
Release factor binds to the stop condon (UAA) to terminate translation and release the complete polypeptide.
Question 88.
How would lac operon operate in E. coli growing in a culture medium, where lactose is present as source of sugar? (All India 2014C)
Answer:
When lactose is present in a medium having E. coli, it will act as a substrate for enzyme 3-galactosidase and switches on the operon.
Hence, it is also termed as an inducer. It inactivates repressor by binding to it and allows RNA polymerase access to the promoter.
Question 89.
Where does peptide bond formation occur in a bacterial ribosome and how? (Foreign 2014)
Answer:
A peptide bond is formed between carboxyl group (-COOH) of amino acid at P-site and amino group (-NH) of amino acid at A-site. It is formed by the enzyme peptidyl transferase in a bacterial ribosome.
Question 90.
(i) Name the scientist who suggested that the genetic code should he made of a combination of three nucleotides.
(ii) Explain the basis on which he arrived at this conclusion. (Delhi 2014)
Answer:
(i) George Gamow suggested that the genetic code should be made of a combination of three nucleotides.
(ii) George stated that a codon must be of three bases in order to code for 20 amino acids, since there are only four bases (i.e. 43 or 4 × 4 × 4 = 64) which code for 20 amino acids.
Question 91.
Why is charging of tRNA necessary during translation process? (All India 2014C)
Answer:
Amino acids are activated in the presence of ATP and linked to their cognate tRNA. This is called charging of tRNA (aminoacylation). The process is required as the formation of peptide bond between the amino acids is favoured energetically, when they are brought together. Activation of amino acids by ATP provides the energy for the formation of peptide bond.
Question 92.
One of the salient features of the genetic code is that it is nearly universal from bacteria to humans. Mention two exceptions to this rule. Why are some codes said to be degenerated? (Foreign 2014)
Answer:
Genetic code is nearly universal except in mitochondrial codons and in some protozoans. Some amino acids are coded by more than one codon, hence some codes are said to be degenerated.
Question 93.
Draw a schematic diagram of lac operon in its switched off position. Label the following
(i) Structural genes
(ii) Repressor bound to its correct positions
(iii) Promoter gene
(iv) Regulatory gene (Foreign 2012)
Answer:
Question 94.
Write the full form of VNTR. How is VNTR different from Probe? (All India 2011)
Answer:
VNTR-Variable Number Tandem Repeat.
Difference between VNTR and Probe is as follows
VNTR:
It is a class of satellite DNA, where a small sequence is arranged tandemly in many copy numbers.
Probe:
It is a radiolabelled VNTR, used for hybridisation with DNA segments in question.
Question 95.
(i) Differentiate between unambiguous and degenerate codons.
(ii) Write two functions of the codon AUG. (All India 2010C)
Answer:
(i) Differences between unambiguous and degenerate codons are as follows
| Unambiguous | Degenerate |
| No ambiguity for a particular codon. For example, GGA is an ambiguous codon, it codes for glycine as well as glutamic acid. | Code is degenerate for a particular amino acid. |
| A particular codon will always code for the same amino acid, where it is found. | One amino acid is coded by more than one codon, e.g. phenylalanine has two codons, i.e. UUU and UUC. |
(ii) Refer to Answer No. 2.
Question 96.
‘A very small sample of tissue or even a drop of blood can help determine paternity’. Provide a scientific explanation to substantiate how it is possible. (All India 2019)
Answer:
DNA fingerprinting is the basis of paternity testing in case of disputes. This technique is used to distinguish between individual of same species by using their DNA sample. The DNA is isolated from the cells and further amplified to produce many copies by using polymerase chain reaction. This amplified DNA is further processed to detect the presence of similarities between the parent and child.
Because, the DNA from sample can be amplified to produce many copies in DNA fingerprinting, only a very small sample of tissue or even a drop of blood can help determine paternity.
Question 97.
Expand ‘BAC’ and ‘YAC’ what are they and what is the purpose for which they are used? (All India 2019,13)
Answer:
BAC — Bacterial Artificial Chromosome
YAC — Yeast Artificial Chromosome.
‘BAC’ and ‘YAC’ are used as vectors in cloning of DNA.
Question 98.
(i) Expand VNTR and describe its role in DNA fingerprinting.
(ii) List any two applications of DNA fingerprinting technique. (2018)
Answer:
(i) VNTR The expanded form of VNTR is Variable Number of Tandem Repeats. These are short nucleotide repeats in the DNA. These are highly specific for individuals. No two individuals have the same VNTR.
Role of VNTR in Fingerprinting VNTRs are used as probe markers in the identification of DNA of different individuals because no two individuals can have the same VNTRs (except in case of monozygotic twins).
(ii) Applications of DNA Fingerprinting:
- DNA fingerprinting can identify the real genetic mother, father and offspring.
- DNA fingerprinting is very useful in the detection of crime and legal pursuits.
Question 99.
(i) List the two methodologies which were involved in human genome project. Mention how they were used.
(ii) Expand YAG and mention what was it used for. (All India 2017)
Answer:
(i) Two major methodologies involved in HGP are as follows
- Expressed Sequence Tags (ESTs) This method focuses on identifying all the genes that are expressed as RNA.
- Sequence annotation This method involves the sequencing of whole set of genome (that contained all coding and non-coding sequence) and then assigning functions to the different regions in the sequence.
(ii) YAC stands for yeast artificial chromosome. These were used as vectors in which fragments of whole DNA of human were inserted and cloned.
Question 100.
A number of passengers were severely burnt beyond recognition during a train accident. Name and describe a modern technique that can help handover the dead to their relatives. (Delhi 2017)
Or
Following the collision of two trains a large number of passengers are killed. A majority of them are beyond recognition. Authorities want to handover the dead to their relatives. Name a modern scientific method and write the procedure that would help in the identification of kinship. (Delhi 2015)
Or
Following a severe accident, many charred-disfigured bodies are recovered from the site making the identification of the dead very difficult. Name and explain the technique that would help the authorities to establish the identity of the dead to be able to handover the dead to their respective relatives. (All India 2014C)
Or
In a maternity clinic, for some reasons the authorities are not able to handover the two newborns to their respective real parents. Name and describe the technique that you would suggest to sort out the matter. (All India 2013)
Answer:
The technique that can help in the identification of victims is DNA fingerprinting which is used to distinguish between individuals of same species by using their DNA sample. The chemical structure of DNA is same in everyone (99.9%) except the order of base pairs, i.e. only 0.1% of DNA makes every individual unique.
DNA fingerprinting exploits the highly variable tandem repeating sequences, i.e. VNTRs for profiling. These VNTRs are highly conserved among members of the same species.
Technique:
This technique is carried out in following steps:
- DNA Isolation DNA is extracted from the cells in a high speed centrifuge.
- Amplification Many copies of the extracted DNA can be made by the use of polymerase chain reaction.
- Digestion of DNA by restriction endonucleases.
- Separation of DNA fragments by electrophoresis.
- Blotting Transfer of separated DNA fragments to synthetic membranes (like nylon or nitrocellulose).
- Hybridisation, with the help of a radio-labelled VNTR probe (small segments of DNA which help to detect the presence of a gene in a long DNA sequence). These probes target a specific nucleotide sequence that is complementary to them.
- Autoradiography Detection of hybridised DNA fragments by autoradiography.
The presence of similarities between the victims and their relatives determines their association on the basis of which dead bodies or newborns can be identified and handed over to their families.
Question 101.
(i) What do ‘Y’ and ‘B’ stand for in ‘YAC’ and ‘BAG used in Human Genome . Project (HGP)? Mention their role in the project.
(ii) Write the percentage of the total human genome that codes for proteins and the percentage of discovered genes whose functions are known as observed during HGP.
(iii) Expand SNPs identified by scientists in HGP. (All India 2016)
Answer:
(i) Y stands for yeast in the word YAC (Yeast Artificial Chromosome) and B stands for bacteria in the word BAC (Bacterial Artificial Chromosome). These are used as vectors in cloning of DNA.
(ii) Less than 2% of the total human genome codes for protein, functions of 50 % of discovered genes are not known.
(iii) SNPs stands for single nucleotide polymorphisms.
Question 102.
(i) How many codons code for amino acids and how many are unable to do so?
(ii) Why are codes said to be
(a) degenerate and
(b) unambiguous? (Delhi 2016C)
Or
(i) How many codons code for amino acids and how many do not?
(ii) Explain the following giving one example of each.
(a) Unamhiguous and specific codon
(b) Degeneration codon
(c) Universal codon
(d) Initiator codon (All India 2010C)
Answer:
(i) Out of 64 codons, 61 code for amino acids and rest 3 codons do not code for any amino acids. These function as stop codons.
(ii) (a) Unambiguous and specific codon These code for only one amino acid, thus, making the genetic code unambiguous and specific, e.g. UUU.
(b) Some amino acids are coded by more than one codon, so the code is degenerate, e.g. serine is coded by 6 codons.
(c) Codon is nearly universal. Some exceptions to the rule are mitochondrial codon and in some protozoans, e.g. UUU.
(d) Initiator codon AUG has dual function. It codes for methionine and also acts as initiator.
Question 103.
Write any three goals of human genome project. (Outside Delhi 2016C)
Answer:
The following are the goals of the human genome project:
- Identify all the (approximately) 20000-25000 genes in human DNA.
- Determine the sequence of the 8 billion chemical base pairs that make up human DNA.
- Improve tools for data analysis.
Question 104.
What is mutation? Explain with the help of an example how does a point mutation affect the genetic code. Name another type of mutation. (Outside Delhi 2016C)
Answer:
A mutation is a sudden, stable, inheritable change in genetic material. A classical example of point mutation is a change of single base pair in the gene for p-globin chain that results in the change of amino acid residue glutamate to valine in a cell of a person suffering from sickle-cell anaemia. It is a genetic disease.
The other type of mutation includes frameshift insertion or deletion mutation. The insertion or deletion of one or more bases may change the reading frame from the point of insertion or deletion. Insertion or deletion of three or its multiple bases insert or delete one or multiple codon, hence one or multiple amino acid may be formed in polypeptide.
Question 105.
Explain the significance of satellite DNA in DNA fingerprinting technique. (All India 2018)
Answer:
Significance of satellite DNA in DNA fingerprinting A DNA satellite is a region that consists of short DNA sequences repeated many times. The variation between individuals in the lengths of their DNA satellites forms the basis of DNA fingerprinting.
DNA satellites are of two types, i.e. microsatellites and minisatellites. Their characteristic that makes them useful for identification is that they are highly polymorphic. The length of each satellite in DNA is inherited.
The length of satellite regions is highly variable between people. These form small peaks during density gradient centrifugation and thus, are invaluable for identification purposes.
Question 106.
Describe how the lac operon operates, both in the presence and the absence of an inducer in E. coli. (All India 2014)
Answer:
Lac operon is made up of one regulatory gene i and three structural genes (z, y, a).
Its function in the presence and the absence of inducer is as follows
(i) When inducer (lactose) is absent, i gene regulates and produces repressor mRNA. The repressor protein binds to the operator region of operon and as a result prevents RNA polymerase to bind to the operon. The operon is switched off in this situation.
(ii) When inducer (lactose) is present, lactose acts as an inducer and binds to the repressor. Thus, forming an inactive repressor. The repressor fails to bind the operator region. The RNA polymerase binds to the operator and transcripts lac mRNA.
Lac mRNA is known to be polycistronic which produces all three enzymes, i.e. P-galactosidase, permease and transacetylase required for the hydrolysis of lact >se. Operon is switched on in this situation.
Question 107.
(i) What is a genetic code?
(ii) Explain the following Degenerate code, Unambiguous code, Universal code, Initiator code. (All India 2014C)
Or
State the conditions when genetic code is said to be
(i) degenerate
(ii) unambiguous and specific
(iii) universal (Foreign 2012)
Or
Unambiguous, universal and degenerate are some of the terms used for the genetic code. Explain the salient features of each of them. All India 2011
Answer:
(i) Genetic code is the sequence of three nucleotides present on tnRNA which codes for a specific amino acid.
(ii) Refer to Answer No. 30 (ii).
Question 108.
Explain the process of translation. (All India 2014C)
Answer:
Translation is the process of polymerisation of amino acids to form a polypeptide with the help of mRNA, tRNA and ribosomes and many enzymes involved in the process.
The different phases of translation are:
- Activation of amino acids (Refer to text on page no. 157).
- Initiation of polypeptide synthesis (Refer to text on page no. 158).
- Elongation of polypeptide chain (Refer to text on page no. 158).
- Termi’nation of polypeptide synthesis (Refer to text on page no. 158).
Question 109.
(i) Which human chromosome has
(a) maximum number of genes and
(b) which one has fewest genes?
(ii) Write the scientific importance of single nucleotide polymorphism identified in human genome. (All India 2014C)
Answer:
(i) Refer to Answer No. 6.
(ii) Scientists have identified about 1.4 million locations, where single base DNA differences or Single Nucleotide Polymorphisms (SNPs) occur in humans. Since, these sequences have high degree of polymorphism they form the basis of DNA fingerprinting.
Question 110.
(i) Name the scientist who postulated the presence of an adapter molecule that can assist in protein synthesis, (ii) Describe its structure with the help of a diagram. Mention its role in protein synthesis. (Foreign 2014)
Answer:
(i) Francis Crick proposed the presence of an adapter molecule, i.e. /RNA which could read the code and bind to the specific amino acids, thus assisting in protein synthesis.
(ii) A clover leaf structure of tRNA
A tRNA- functions as carrier of amino acids and participates in protein synthesis.
Question 111.
(i) Explain DNA polymorphism as the basis of genetic mapping of human genome.
(ii) State the role of VNTR in DNA fingerprinting. (All India 2013)
Answer:
(i) DNA polymorphism exhibited by certain repetitive DNA sequences is used to construct genetic and physical maps of the genome which are used in human genome project. (ii) Variable Number of Tandem Repeats (VNTRs) belong to a class of satellite DNA called as minisatellite. VNTRs are used as probes in DNA fingerprinting.
Question 112.
Given below are the sequences of nucleoside in a particular mRNA and amino acids coded by it.
UUUAU GUUC GAGUUAGU GUAA
Phe – Met – Phe – Glu – Leu – Val
Write the properties of genetic codes that can be and that cannot be correlated from the above given data. (Delhi 2013C, 2010C)
Answer:
UUUAUGUUCGAGUUAGUGUAA
Phe – Met – Phe – Glu – Leu – Val
According to the sequence given above
- Codon is triplet.
- Genetic code is specific and unambiguous.
For example, AUG – Codes for methionine (Met) GAG – Codes for glutamine (Glu) UUA – Codes for leucine (Leu) - Codon is degenerate, i.e. same amino acids are coded by more than one code.
For example, UUU and UUC, both codes for phenylalanine (Phe). - Code is read without punctuation.
- UAA acts as a terminating code.
Thus, all properties of codon are satisfied from the above given data. Except the one that mostly AUG work as an initiating codon.
Question 113.
How are the structural genes activated in lac operon in E. coli ? (All India 2012)
Answer:
Lac operon consists of
- an operator, which controls all structural genes as a unit
- a regulatory gene (i gene)
- three structural genes (z, y, a), which code for enzymes and
- a promoter, where RNA polymerase binds for transcription.
The regulatory gene codes for a repressor protein that has high affinity for the operator region and which prevents RNA polymerase from transcribing the structural genes, i.e. the lac operon is switched off or inactive.
Lac operon in the presence of inducer When inducer (lactose) is added, it binds with repressor protein and inactivates it. This allows RNA polymerase to have access to the promoter and transcription proceeds via the activation of structural genes.
Question 114.
(i) Name the scientist who called tRNA an adapter molecule.
(ii) Draw a clover leaf structure of tRNA showing the following
(a) Tyrosine attached to its amino acid site.
(b) Anticodon for this amino acid in its correct site (codon for tyrosine is UAC).
(iii) What does the actual structure of tRNA look like? (All India 2011)
Answer:
(i) Francis Crick.
(ii) Refer to Answer No. 38 (ii).
(iii) The actual structure of tRNA looks like an inverted L.
Question 115.
A considerable amount of lactose is added to the growth medium of E. coli. How is the lac operon switched on in the bacteria? Mention the state of the operon when lactose is digested. (All India 2010C)
Answer:
In lac operon, when lactose is added, it enters the cell wall with the help of permease, a small amount of which is already present in cell. Lactose binds itself to active repressor and changes its structure. The repressor now fails to bind to the operator.
Then, RNA polymerase starts transcription of operon by binding to promoter site-P. All the three enzymes for lactose metabolism are synthesised.
After some time, when whole of lactose is consumed, there is no inducer present to bind to the repressor. Due to this, the repressor becomes active again, attaches itself to the operator and finally switches off the operon.
Question 116.
(i) Why is tRNA called an adapter?
(ii) Draw and label a secondary structure of tRNA. How does the actual structure of tRNA look like? (All India 2010C)
Answer:
(i) tRNA binds to a specific amino acid and it also reads the codon of the amino acid bound to it through its anticodon. So, it is called an adapter molecule.
In actual structure, the /RNA is a compact molecule, that looks like an inverted L.
Question 117.
The following is the flow chart higlighting the steps in DNA fingerprinting technique. Identify a, b, c, d, e and f.
Answer:
a – Restriction endonuclease
b – Polyacrylamide gel
c – Nitrocellulose or Nylon
d – VNTR e – Hybridisation
e- Autoradiography
Question 118.
Study the schematic representation of the genes involved in the lac operon given below and answer the questions that follows
(i) Identify and name the regulatory gene in this operon. Explain its role in switching off the operon.
(ii) Why is lac operon’s regulation referred to as negative regulation?
(iii) Name the inducer molecule and the products of the genes z and y of the operon. Write the function of these gene products. (All India 2019, Foreign 2010, Delhi 2010)
Answer:
(i) i gene-regulatory gene : It codes for the repressor protein of the operon, which is synthesised constitutively. The repressor has the affinity for the operator gene. It binds to the operator and prevents the RNA polymerase from transcribing the structural genes.
(ii) When repressor binds to the operator, the operon is switched off and transcription is stopped. So, it is called negative regulation.
(iii) Lactose is an inducer molecule.
Gene ‘Z’ codes for p-galactosidase, which is responsible for the hydrolysis of lactose into galactose and glucose.
‘y’ gene codes for permease. It increases the permeability of the cell to lactose.
Question 119.
(i) Write the contributions of the following scientists in deciphering the genetic code.
George Gamow, Har Gobind Khorana, Marshall Nirenberg, Severo Ochoa.
(ii) State the importance of genetic code in protein biosynthesis. (Delhi 2019)
Answer:
(i) George Gamow coined the term genetic code and argued that since there are only four bases and if they have to code for 20 amino acids, the code should constitute a contribution of bases.
He suggested that in order to code for all* the amino acids the genetic code should be made up of 3 nucleotides.
Har Gobind Khorana developed a chemical 52 method for the synthesis of RNA molecule with defined base combinations (homopolymers and copolymers). Marshall Nirenberg Put forward a cell-free system for protein synthesis that helps in deciphering the code.
Severo Ochoa Showed that the polynucleotide phosphorylase also helped in polymerising RNA with defined sequences in a template independent manner (enzymatic RNA synthesis).
(ii) The genetic code is the set of rules by which information encoded in genetic material is translated into proteins by living cells. The genetic code is nearly universal language that encodes directions for cells. Their arrangement as codons, stores the blueprint for amino acid chain. This chain in turn forms proteins which regulate the biological process in every living beings.
Question 120.
What is an operon? Explain the functioning of operon when in an open state. (2018C)
Answer:
For operon and functioning of operon, Refer to text on page no. 159 and 160.
Question 121.
(i) Explain the role of regulatory gene, operator, promoter and structural genes in lac operon when E. coli is growing in a culture medium with the source of energy as lactose.
(ii) Mention what would happen if lactose is withdrawn from the culture medium. (Delhi 2016C)
Answer:
(i) For role of regulatory gene, operator gene, promoter gene and structural gene, Refer to text on page no. 159 and 160.
(ii) If lactose is withdrawn from the culture medium, the repressor of the operon is synthesised from the i gene. This repressor protein binds to the operator region of operon and prevents RNA polymerase from transcribing the operon.
Question 122.
(i) Write any two different levels at which regulation of gene expression could be exerted in eukaryotes.
(ii) Give a labelled schematic representation of ‘lac operon’ in its switched off position. (All India 2016)
Answer:
(i) The different levels at which regulation of gene expression can be exerted in eukaryotes are following
- Transcriptional level (formation of primary transcript).
- Processing level (regulation of splicing)
(ii) Lac operon in switched off position
Refer to Answer No. 21.
Question 123.
(i) How is DNA fingerprinting done? Name any two types of human samples which can be used for DNA fingerprinting. Explain the process sequentially.
(ii) Mention any two situations when the technique is useful. Outside Delhi 2016C
Answer:
(i) Refer to Answer No. 28.
Samples that can be used for DNA fingerprinting are nails, blood, hair, skin, etc.
(ii) The technique is useful in the following situations
- Paternity testing in case of disputes.
- In determining population and genetic diversities.
Question 124.
How do mRNA, tRNA and ribosomes help in the process of translation? (All India 2015)
Or
Name the major types of RNAs and explain their role in the process of protein synthesis in a prokaryote. (Foreign 2014)
Answer:
(i) Translation is the process of polymerisation of amino acids to form a polypeptide. The role of mRNA, tRNA and ribosomes in the process is described below
(a) Role of mRNA The order and sequence of amino acids are defined by the sequence of bases in the mRNA. It stores the genetic information from DNA. The amino acids are joined by a bond, which is known as a peptide bond.
This process requires energy mRNA also possesses untranslated sequences called Untranslated Regions (UTR) at both 5′ end and 3′ end. They help in efficient translation.
(b) Role of (RNA It acts as an adaptor molecule. Activation of amino acids occurs in the presence of ATP and activated amino acids get linked to their cognate tRNA, i.e. charging of fRNA or aminoacylation of tRNA.
If two such charged fRNAs are brought closer, the formation of peptide bond between them would occur energetically in the presence of a catalyst.
(c) Role of /RNA It is formed in nucleolus and it forms 80% of total RNA present inside the cell. It is also the most stable type of RNA. rRNA is associated with structural organisation of ribosomes (rRNA forms about 60% of weight of ribosomes), which are seats of protein synthesis.
(d) Role of ribosomes Initiation of polypeptide synthesis occurs in ribosomes (cellular factory for protein synthesis).
- Ribosome consists of structural RNAs and about 80 different proteins.
- In its inactive state, it exists as two subunits, i.e. a large and a small subunit.
- When the small subunit encounters an mRNA, the process of translation of the mRNA to protein begins.
- There are two sites in the large subunit, i.e. the P-site and A-site for subsequent amino acids to bind to and thus, be close enough to each other for the formation of a peptide bond.
The small subunit (with the fRNA) attaches to the large subunit in such a way that the initiation codon (AUG) comes to the P-site.
Question 125.
Sketch a schematic diagram of lac operon in switched on position. How is the operon switched off ? Explain. (Outside Delhi 2015C)
Answer:
(i) Refer to Answer No. 2.
Question 126.
(i) Write the specific features of the genetic code AUG.
(ii) Genetic codes can be universal and degenerate. Write about them, giving one example of each.
(iii) Explain aminoacylation of the tRNA. (All India 2013)
Answer:
(i) Refer to Answer No. 2.
(ii) Refer to Answer No. 30 (ii).
(iii) Refer to Answer No. 14.
Question 127.
Given below is the schematic representation of lac operon of E.coli. Explain the functioning of this operon when lactose is provided in the growth medium of the bacteria.
Answer:
Lactose in the lac operon regulates operon, by switching on and off. If lactose is present in the medium, it acts as an inducer here and the substrate for the enzyme p-galactosidase.
It binds to the repressor and forms an inactive repressor, which fails to bind to the operator region of the operon.
The RNA polymerase thus, binds to the operator and transcribes lac-mKNA. Lac mRNA produces all three enzymes, i.e. P-galactosidase, permease and transacetylase of polycistronic structural gene present in E.coli.
Therefore, operon will be switched on in the presence of the lactose.
Question 128.
(i) DNA polymorphism is the basis of DNA fingerprinting technique. Explain.
(ii) Mention the causes of DNA polymorphism. (Foreign 2010)
Answer:
(i) DNA polymorphism It is the occurrence of inheritable mutations at a frequency greater than 0.01 in a population.
- Such variations often occur in non-coding sequences. They keep on accumulating generation after generation.
- Types of polymorphism range from single nucleotide change to very large scale changes.
- Single nucleotide polymorphism is used to diagnose disease related sequences of DNA on the chromosome.
- Variable number of tandem repeats show a high degree of polymorphism.
(ii) DNA polymorphism occurs due to mutations.
Refer to text on page no. 161-162, for further detail.
Question 129.
Name and describe the technique that will help in solving a case of paternity dispute over the custody of a child by two different families. (All India 2010)
Answer:
Refer to Answer No. 28.
Question 130.
(i) Name the enzyme responsible for transcription of tRNA and the amino acid, the initiator tRNA gets linked with.
(ii) Explain the role of initiator tRNA in initiation of protein synthesis. (Delhi 2012)
Answer:
(i) RNA polymerase transcribes tRNA and the amino acid, the initiator tRNA gets linked with is methionine.
(ii) The initiator tRNA binds the amino acid methionine at its amino acid acceptor site. It has anticodon loop, which has anticodon for methionine, i.e. UAC. It recognises the start codon (AUG) at P-site and binds to it according to complementarity of bases.
Question 131.
(i) Describe the process of aminoacylation.
(ii) ‘Process of transcription and translation are coupled in prokaryotes, but not in eukaryotes’. Explain. (All India 2019)
Answer:
(i) Refer to Answer No. 14 on page no. 171.
(ii) In bacteria, both processes occur in cytoplasm as there is no nucleus. In eukaryotes, transcription occurs in nucleus, while translation occurs in the cytoplasm.
Complexities in eukaryotic transcription: Eukaryotes have additional complexities in gene expression than prokaryotes as mentioned in gene expression below
(a) There are at least three RNA polymerases in the nucleus other than the RNA polymerase in organelles. The RNA polymerase-I transcribes rRNAs (28S, 18S and 5.8S). RNA polymerase-III is responsible for the transcription of tRNA, 5srRNA and snRNAs (small nuclear RNAs). RNA polymerase-II transcribes precursor of mRNA, the heterogeneous nuclear RNA (hnRNA).
(b) Another complexity is that, the primary transcripts contain both the exons and the introns and are nonfunctional. Hence, it is subjected to a process called splicing.
In this process, introns are removed and exons are joined in a definite order.
(c) hnRNA undergoes additional processing called capping and tailing. In capping, an unusual nucleotide is added to the 5′ end of ImRNA.
In tailing, adenylate residues (200-300) are added at 3′ end in a template. Capping and tailing protect the mRNA from degradation by the activity of digestive enzymes present in the cytoplasm. It is the fully processed hnRNA, now called mRNA, that is transported out of the nucleus for translation process.
Complexities in translation in eukaryotes
- The mRNA formed in nucleus has to be transported to the cytoplasm.
- Transcription and translation cannot be coupled in eukaryotes.
Question 132.
(i) Describe the structure and function of a tRNA molecule. Why is it referred to as an adapter molecule?
(ii) Explain the process of splicing of hnRNA in a eukaryotic cell. (All India 2017)
Answer:
(i) Structure of tRNA
- The secondary structure of tRNA looks like a clover-leaf.
- All tRNA molecules have a guanine residue at its 5’end.
- At its 3′ end an unpaired CCA sequence is present. Amino acids get attached to this end during translation.
- tRNA has an anticodon loop, an amino acid acceptor arm, and a ribosome site.
- The anticodon loop has bases complementary to the code. Amino acid acceptor end binds to amino acids.
Function of tRNA The function of tRNA is to align the required amino acids according to the nucleotide sequence of mRNA. tRNA is also called the adapter molecule because on one hand it can read the code and on the other hand it can bind to specific amino acid. It acts as intermediate molecule between triplet code of mRNA and amino acid sequence of polypeptide chain.
(ii) Splicing of hnRNA Eukaryotic transcripts possess extra segments called introns or intervening sequences or non-coding sequences. They do not appear in mature or processed RNA. The functional coding sequences are called exons. Splicing is removal of introns and fusion of exons to form functional RNAs. A complex called spliceosome is formed between 5’end (GU) and 3’end (AG) of intron. Energy is obtained from ATP. It removes the introns. The adjacent exons are brought together. The ends are sealed by RNA ligase.
Question 133.
Where do transcription and translation occur in bacteria and eukaryotes, respectively? Explain the complexities in transcription and translation in eukaryotes that are not seen in bacteria. (Foreign 2010)
Answer:
In bacteria, both processes occur in cytoplasm as there is no nucleus.
In eukaryotes, transcription occurs in nucleus, while translation occurs in the cytoplasm. Complexities in eukaryotic transcription Refer to Answer No. 2. (ii)
Complexities in translation in eukaryotes are
- The OTRNA formed in nucleus has to be transported to the cytoplasm.
- Transcription and translation cannot be coupled in eukaryotes.
Question 134.
According to human genome project, about 99.9% nucleotide bases are exactly the same in all humans. Do you think the discrimination of people on the basis of colour, creed and religion is correct? Justify.
Answer:
No discrimination of people on these grounds is not correct because all people have same genetic material (i.e. DNA) and are similar in their makeup.
Question 15.
An organism is able to survive on a culture medium, containing nutrient A, by the enzyme-catalysed reactions.
A mutant organism failed to survive on this medium, but grew well when nutrient C was added to it.
(i) Which gene of this mutant organism is defective?
(ii) What does such a condition indicate of?
(iii) Indicate the value expressed in this sequence of reactions.
Answer:
(i) The gene q is defective.
(ii) It indicates that one gene controls the synthesis of one enzyme.
(iii) Each one of reaction has a role to play, when one fails, progress is not possible.










